
Privacy statement: Your privacy is very important to Us. Our company promises not to disclose your personal information to any external company with out your explicit permission.
Select Language
Nickel ion metal chelating affinity chromatography medium (Ni-NTA) specification
one, Introduction
Metal chelate affinity chromatography medium, also known as fixed metal ion affinity chromatography, is based on the use of some amino acids on the surface of the protein, such as histidine and various transition metal ions such as Cu 2+ , Zn 2+ , Ni 2 + , Co 2+ , Fe 3+ have a special interaction, which can adsorb proteins rich in such amino acids, thus achieving the purpose of separation and purification. Therefore, an agarose gel coupled with these metal ions can selectively separate these proteins containing a plurality of histidines and polypeptides, proteins and nucleotides which adsorb metal ions. Cysteine and tryptophan can also bind to immobilized metal ions, but this binding force is much less than the binding of histidine residues to metal ions.
Nickel NTA affinity chromatography medium (Ni-NTA) has the advantages of good specificity and fast flow rate, uniform particle size, small particle size, and more stable chelated nickel, which can withstand higher reducing agents, physical and chemical stability. Good sex, good batch repeatability. This product has chelated nickel ions and is more convenient to use.
Second, the performance parameters:
Characteristics | High density of group, large load, high resolution and easy to use |
Matrix | 6% crosslinked agarose gel |
Ligand | Ni 2+ |
Ligand density | 20 - 40μmol / ml |
Adsorption capacity | 15mg protein / ml |
Media particle size | 45 - 165μm |
Maximum flow rate | 600cm/h |
pH range | 3 - 10, pH range up to 2 - 11 during in-place cleaning |
Storage temperature | +4 - 8°C |
Preserving liquid | 20% ethanol |
Third, the scope of application
A His- tagged recombinant protein and a polypeptide, protein, nucleotide, and phosphorylated protein capable of being adsorbed by a metal ion are separated .
Fourth, application examples
Experimental name: Ni-NTA Separation of His- tagged recombinant protein
Experimental steps:
1. Ni-NTA packed column, 1.6×20cm, and the bed volume is 10ml;
2. Balance 2 to 5 bed volumes with buffer 1 at a flow rate of 2 ml/min;
3, 20ml cell disruption solution (50mM PBS, pH 7.4, 0.5M NaCl) 0.45μm filter membrane filtration, loading, flow rate of 1ml / min;
4, wash 2 to 5 bed volume with buffer 1 , the flow rate is 2ml / min;
5. Stage elution with buffer 3 containing 10, 20, 50, 100, 200, 300, 400 mM imidazole at a flow rate of 2 ml/min, collect the elution peaks at each stage, and measure the molecular weight of the fusion protein by SDS-PAGE. Size and purity;
6. Wash 5 bed volumes with pure water, then wash 3 bed volumes with 20% ethanol, flow rate is 2ml/min, and the column is stored in +4 - 8 °C environment.
Buffer composition:
Buffer 1 : 50 mM PBS buffer pH 7.4. Formulation: 0.5 M NaH 2 PO 4 19 ml, 0.5 M Na 2 HPO 4 81 ml, NaCl 29.3 g, dissolved in an appropriate amount of water to a volume of 1000 ml.
Buffer 2 : 50 mM phosphate buffer, pH 7.4, a PBS solution of pH 7.4. Formulation: 0.5 M NaH 2 PO 4 19 ml, 0.5 M Na 2 HPO 4 81 ml, NaCl 29.3 g and imidazole 34 g, add appropriate amount, adjust the pH to 1000 ml after adjusting the pH with water.
Buffer 3 : Buffer B with different imidazole concentrations:
Imidazole concentration | Buffer 1 amount (ml) | Buffer 2 amount (ml) |
10 mM | 98 | 2 |
20 mM | 96 | 4 |
50 mM | 90 | 10 |
100 mM | 80 | 20 |
200 mM | 60 | 40 |
300mM | 40 | 60 |
400 mM | 20 | 80 |
SDS-PAGE process:
1, BCA method to measure sample protein concentration
2. Calculate the volume required for 5-10 μg/well based on the protein concentration of the assay sample.
3. Add a sample solution containing 5-10 μg of protein to a 1.5 ml EP tube. If the volume is less than 10 μl, add 10 μl of 20 mM PBS pH 7.4; if the volume is larger than 10 μl, add 1 ml of absolute ethanol at -20 ° C. Concentrated for 1 h.
4. Centrifuge the concentrated sample at 10,000 rpm for 15 min, remove the supernatant, and remove the residual ethanol in an oven at 37 ° C for 10 min.
5. Add 10 μl of each of 20 mM PBS pH 7.4 and 2×loading buffer to the sample for 10 min at 100 °C. After taking out, it was cooled for 30 s and centrifuged at 4000 rpm for 1 s.
6, spotting, electrophoresis.
Experimental results:
(1) Purification of His- tag recombinant protein using Ni-NTA Agarose
The amount of His-tag recombinant protein was 20 ml, and eluted with buffer B containing 20, 50, 100, 200, 300, 400 mM imidazole, respectively. The chromatographic results are shown in Figure 1. For the SDS-PAGE results of the chromatographic components, see figure 2.
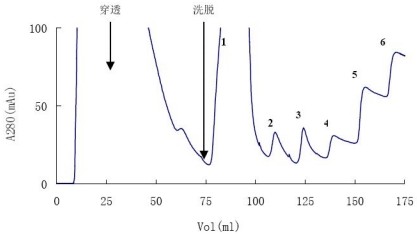
Figure 1. Chromatogram of Ni-NTA Agarose Purified His- tag Recombinant Protein

Figure 2. SDS-PAGE map
1, Standard protein, 2, sample solution, 3. flow through, 4:20 mM eluate, 5:50 mM eluate,
6: 100 mM eluate, 7:200 mM eluate, 8:300 mM eluent, 9:400 mM eluate.
(2) Purification of His- tag recombinant protein inclusion bodies using Ni-NTA Agarose
The conditions are basically the same as the previous method, except that 8M urea is added to the buffer. The solution formula is as follows. The chromatogram and electrophoresis map are shown in Fig. 3 and Fig. 4.
It should be noted that different inclusion bodies have different solubility. It is also possible to replace 8M urea with 6M guanidine hydrochloride because guanidine hydrochloride dissolves inclusion bodies more completely.
Buffer composition:
Buffer 1 : 50 mM PBS buffer pH 7.4. Formulation: 0.5 M NaH 2 PO 4 19 ml, 0.5 M Na 2 HPO 4 81 ml, NaCl 29.3 g and urea 480 g, dissolved by heating to a volume of 1000 ml.
Buffer 2 : 50 mM pH 7.4 in PBS. Formulation: 0.5M NaH 2 PO 4 19ml, 0.5M Na 2 HPO 4 81ml,
NaCl 29.3 g, imidazole 34 g and urea 480 g, dissolved in heat to a volume of 1 000 ml.
Buffer 3 : Buffer B with different imidazole concentrations:
Imidazole concentration | Buffer 1 amount (ml) | Buffer 2 amount (ml) |
50 mM | 90 | 10 |
400 mM | 20 | 80 |

FIG 3. Ni-NTA Agarose was purified His-tagged recombinant protein inclusion bodies chromatograms

FIG 4. SDS-PAGE in FIG.
1: inclusion body, 2: flowthrough liquid, 3: 50mM imidazole eluate, 4: 400mM imidazole eluate.
(3) Effect of different metal ions and elution conditions on purification
Using Ni-NTA Agarose, the His tag recombinant protein was loaded in 10 ml, eluted with buffer 3 containing 20, 50, 100, 200, 500 mM imidazole, and buffers 1 and 2 were added to a final concentration of 1%. At a temperature of 80, the results show that the addition of a surfactant can reduce the adsorption of impurities. In addition, the same purification experiments were carried out by chelation of copper-cobalt metal ions. The results showed that the recovery of nickel ions was the best in recovery and purity, and the purity of separation and purification could be >90%, and the recovery rate of target protein was as high. 80%, so it is recommended to use nickel ion chelating fillers, others can be ignored.
Fifth, application notes:
The most classical ligand for nickel ion metal chelate affinity chromatography media is IDA. Nickel ion has six chelating valences, Ni-IDA sequesters trivalent, and residual trivalent; while Ni-NTA chelates tetravalent, leaving two valences, so Ni-IDA agarose gel has a stronger force than Ni- NTA agarose gel is strong. For this reason, under the same conditions, Ni-IDA washes impurities and target proteins higher than the imidazole concentration of Ni-NTA , but the NTA filler is more stable, withstands stronger reducing agents, and is less likely to fall off; IDA 's load is higher than NTA , and it can be used repeatedly and more economically. Which filler is used depends on the individual's habits and the conditions of purification.
Six, the relevant operating instructions
1. Column packing
(1) The temperature of all materials to be used should be the same as the temperature of the chromatographic operation. The liquid is preferably degassed.
(2) Distilled water is added to the lower end of the column to remove the air in the column, and the column outlet is closed, and a small amount of distilled water is retained in the column.
(3) When the medium is continuously poured into the column, the glass rod should be used to drain against the inner wall of the column to reduce the generation of bubbles and allow the medium to settle naturally.
(4) The column pressure does not exceed 0.3 MPa. If the column pressure cannot be measured in the column packing system, the control flow rate is higher than 300 cm/h, but generally only 75% of the maximum flow rate is used in use.
2, fixed metal ions
(1) The metal ion must be fixed with a filtered metal ion solution to prevent precipitation of the metal salt on the medium.
(2) The column is sufficiently equilibrated with 2-5 bed volumes of distilled water.
(3) Select suitable metal ions (Cu 2+ , Zn 2+ , Ni 2+ , Co 2+ , Fe 3+ , etc.) and dissolve them in a neutral or weakly acidic solution at a concentration of 0.1 to 0.3M. If it is Fe 3+ it must be chelated at a low pH (pH 3) to prevent precipitation of Fe 3+ .
(4) The column is eluted with 2-5 bed volumes of metal ion solution, and the column is washed with not less than 5 times the bed volume of distilled water to wash away the unchelated metal ions. (Of course, it is also possible to squirt the washed non-chelating filler directly with the desired metal ion solution on a shaker overnight, so that the chelation effect is better, and Ni-NTA Agarose is particularly suitable for such a method.)
(5) Equilibrate the column with 2 - 5 times the volume of the initial buffer solution, and then load.
(6) If it is chelated iron ions, it should be noted that under neutral conditions, Fe 3+ is easily reduced to form a precipitate, so the pH of the Fe 3+ solution is preferably 3 - 5. Columns that chelate Fe 3+ cannot be stored in neutral solutions for long periods of time. It is recommended that the chelated Fe 3+ be washed with 50 mM EDTA solution after each use and re-chelated the next time it is used. If the wash is not clean, the medium can also be immersed in 50 mM EDTA overnight before being washed and stored.
3, loading
(1) The sample is dissolved in a buffer of pH 5.5 to 8.5, and the pH of the loading buffer is increased to increase the load.
(2) The selection of the starting buffer is mainly based on the characteristics of the metal ion and the binding characteristics of the sample to the metal ion.
(3) The buffer should not contain EDTA and citrate, and it is also preferable to contain no reducing agent such as mercaptoethanol.
(4) Commonly used buffer has 10~20m M sodium phosphate buffer and 50mM sodium acetate buffer
(5) 0.15~0.5M NaCl should be added to the buffer to eliminate ion exchange.
(6) There is a general rule for the use of metal chelate chromatography. If you do not understand the binding properties of the protein, it is recommended to use Zn 2+ first . The buffer can be selected from neutral phosphate or acetate buffer. The NaCl content is 0.15-0.5 M as the starting buffer.
(7) Detergents in buffer generally do not affect the adsorption of proteins.
(8) When the protein is adsorbed, a part of the chelated metal ions are often replaced. This phenomenon is usually visible, especially when using colored metal ions, such as Cu2+, so the metal ions can be used first after several times. Wash off and then re-chelate the metal ions.
4, elution
(1) Linear reduction or one-step lowering of pH, most proteins will be eluted at pH 6~4, or at pH 3-4, and the buffer may be sodium acetate, citric acid or phosphate buffer system.
(2) Competitive elution: a substance that increases linearly or increases affinity with metal ions in a step, such as 0-0.5 M imidazole, 0-50 mM histidine, 0-2 M NH 4 Cl. The gradient elution is preferably carried out at a constant pH of the starting buffer.
(3) Chelating agents such as EDTA and EGTA will react with metal ions, causing the protein to be eluted. This method can not separate different proteins, and it will affect protein adsorption, resulting in the fusion protein not hanging.
(4) In all the above cases, 0.15~0.5M NaCl must be added to the buffer to eliminate ion exchange.
(5) When the chelating ion ligand is Cu 2+ , there are the following three modes of operation:
Lower the pH:
Loading buffer: 50 mM Na 2 HPO 4 , 0.5 M NaCl, pH 7.4
Elution buffer: 50 mM Na 2 HPO 4 , 0.5 M NaCl, pH 3.5
Competitive elution:
Loading buffer: 50 mM Na 2 HPO 4 , 1 M NaCl, pH 7.4
Elution buffer: 50 mM Na 2 HPO 4 , 1M NH 4 Cl, pH 7.4
Shedding and elution:
Loading buffer: 50 mM Na 2 HPO 4 , 0.5 M NaCl, pH 7.4
Elution buffer: 50 mM Na 2 HPO 4 , 0.5 M NaCl, 50 mM EDTA, pH 7.4
The use of reduced pH and exfoliation will cause the metal ions to fall off and re-chelate the metal ions the next time they are used.
Seven , regeneration, cleaning, preservation
1 , the regeneration of the gel
(1 ) The gel must be regenerated before chelation of a new metal ion. The column was rinsed with 5 to 10 volumes of 50 mM EDTA, and the residual EDTA was washed away with 2 to 3 volumes of 0.5 M NaCl.
(2) The method of re-fixing metal ions is as described above. In some operations, denatured proteins and lipids cannot be eluted during the regeneration of the column and they can be removed by in situ cleaning.
2 , in place cleaning
( 1 ) The protein adsorbed by the ion exchange is removed, and the column is rinsed with 2 to 3 times of a bed volume of 2 M NaCl solution, followed by reverse rinsing.
( 2 ) The protein precipitate and hydrophobic protein were removed, and the column was rinsed with 1 M NaOH at a rate of 100 cm/h for 1 h.
( 3 ) In all operations, the column is washed with at least 3 bed volumes of initial buffer.
( 4 ) Remove strong hydrophobic proteins and lipids, wash the column with 4 times bed volume of 70% ethanol or 30% isopropyl alcohol, and then rinse back.
Eight , save
Stored in 20% ethanol for a long time at 4 °C.
September 25, 2024
September 25, 2024
Contactar proveedor
September 25, 2024
September 25, 2024

Privacy statement: Your privacy is very important to Us. Our company promises not to disclose your personal information to any external company with out your explicit permission.

Fill in more information so that we can get in touch with you faster
Privacy statement: Your privacy is very important to Us. Our company promises not to disclose your personal information to any external company with out your explicit permission.